*** Outline ***
Denoising an FID with SVD
Denoising an FID increases the signal to noise ratio of the corresponding spectrum without broadening the linewidth. Our Java applet combined with JCAMP-DX file allows us to apply the SVD denoising method to data acquired with any NMR spectrometer.
Java application
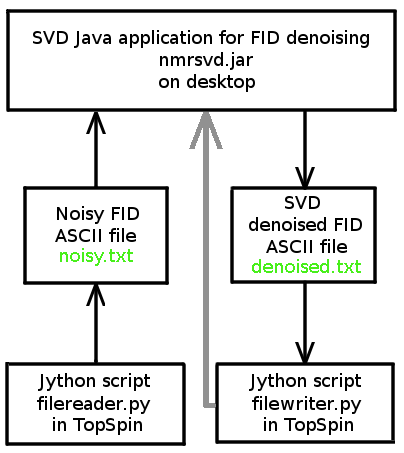
We provide a Java application (nmrsvd.jar) and two Jython scripts for TopSpin (filereader.py and filewriter.py written by Redouane HAJJAR), which allow us to denoise FID signals acquired with Bruker Avance spectrometer. SVD Java application and TopSpin exchange FID data via two ASCII files: noisy.txt and denoised.txt.
The Java application nmrsvd.jar remains valid for other NMR spectrometers. User should have scripts similar to our two jython scripts for data exchange with nmrsvd.jar
Installation
- Download the Java 1.4 application nmrsvd.jar (last updated January 24, 2010) on the desktop of the computer.
- Download and extract the two Jython 2.1 scripts filereader.py and filewriter.py then install them in bruker / topspin / exp / stan / nmr / py / user folder of TopSpin.
- Under Topspin 3.2, replace in the two files the line
filepath = result[3] + "/data/" + result[4] + "/nmr/" + result[0] + "/" + result[1]
by
filepath = result[3] + "/" + result[0] + "/" + result[1]
Execution
In TopSpin:
- Make a copy of experiment data with the wrpa command then display the FID.
- Apply the convdta command if data are obtained in digital mode.
- Provide the processing parameter TDeff with an even integer smaller than 3K, corresponding to the size of the FID for denoising.
- Enter the Jython script filename filereader.py in the command line of TopSpin, which generates the noisy FID ASCII file noisy.txt in the folder of TopSpin containing the FID file.
On desktop:
- Double click the nmrsvd.jar icon to start the program.
- Read the noisy FID ASCII file noisy.txt via File / Open file... menu item, the data are displayed in the two textArea.
- Click Run SVD button for denoising. The duration of calculation depends on the TDeff value and the size of random access memory. The singular values and their graph are shown.
- Provide a reduced number of singular values in the white field, then click Hankel button. The denoised FID is shown in the two textArea.
- Provide the denoised FID ASCII filename denoised.txt in the folder of TopSpin containing the FID file by selecting File / Save as... menu item.
In TopSpin:
- Enter the Jython script filename filewriter.py in the command line of TopSpin, which transforms denoised.txt into FID file.
- Display the denoised FID file in TopSpin, then process the data.
- Jump to Point 3 of "On desktop" part for more denoising if the signal to noise ratio is not good enough.
Reference
- Wikipedia: Endianness.
- Intel: Endianness white paper (PDF file).
- betterexplained: Understanding big and little endian byte order.
- Oracle: ByteOrder (Java platform SE 6).
- mindprod: Endian: Java glossary.
Java stores binary values internally and in files MSB (Most Significant Byte) first, i.e. high order part first. This is referred to as big-endian byte sex or sometimes network order.
- java2s: Endian differences and data storage: Buffering file input output Java.
- darksleep: Java and unsigned types.
- javaworkshop.sourceforge: Java archive (Jar) files.
- Oracle: Packaging programs in JAR files.
- Oracle: The Java Tutorials.
- Carl Albing and Michael Schwarz: Java™ Application Development on Linux (3.4 Mb PDF file).
- Elliotte Rusty Harold: Java I/O.
- jguru: Store and retrieve Unicode or Double byte data in a file using the java.io libraries.
- Kerr, Christopher (2013): Using the Python scripting functionality in Bruker Topspin. (Power Point presentation)
Avance spectrometer with Linux operating system
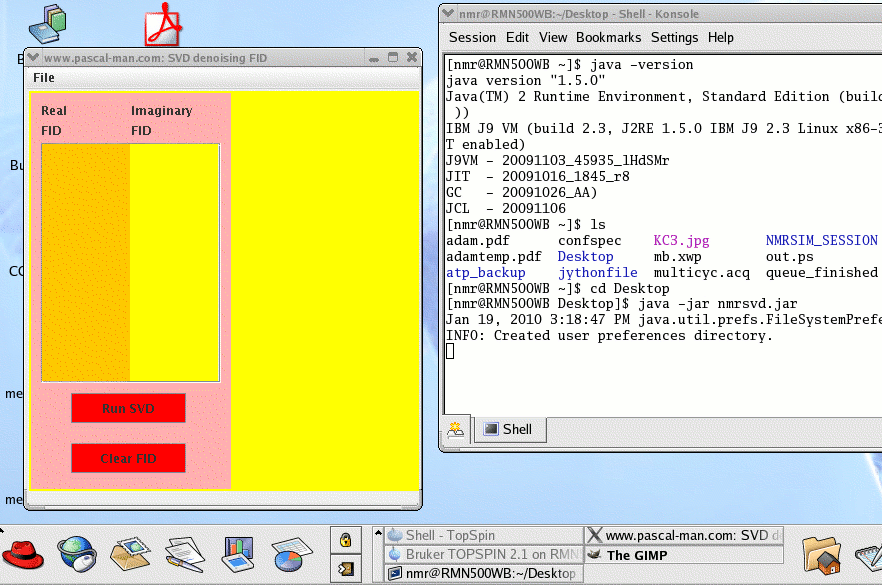
To start nmrsvd.jar in Linux OS, first launch a terminal window (Desktop - Shell - Konsole) called nmr@RMN500WB in our case.
(1) Check the version of Java: java -version
(2) Find the location of Desktop : ls
(3) Move to Desktop directory: cd Desktop
(4) Launch nmrsvd.jar: java -jar nmrsvd.jar